Reaction Model
MetAMDB allows users to upload their own metabolic reaction models. MetAMDB makes it possible for users to generate an atom mapping model for their respective reaction models. The following sections will describe how to create such a reaction model and what options are available.
Download this example model to follow along! (based on Young et al. 2008)
File Format#
Currently, the only available model format is CSV. Other file formats will be available in the future!
File Structure#
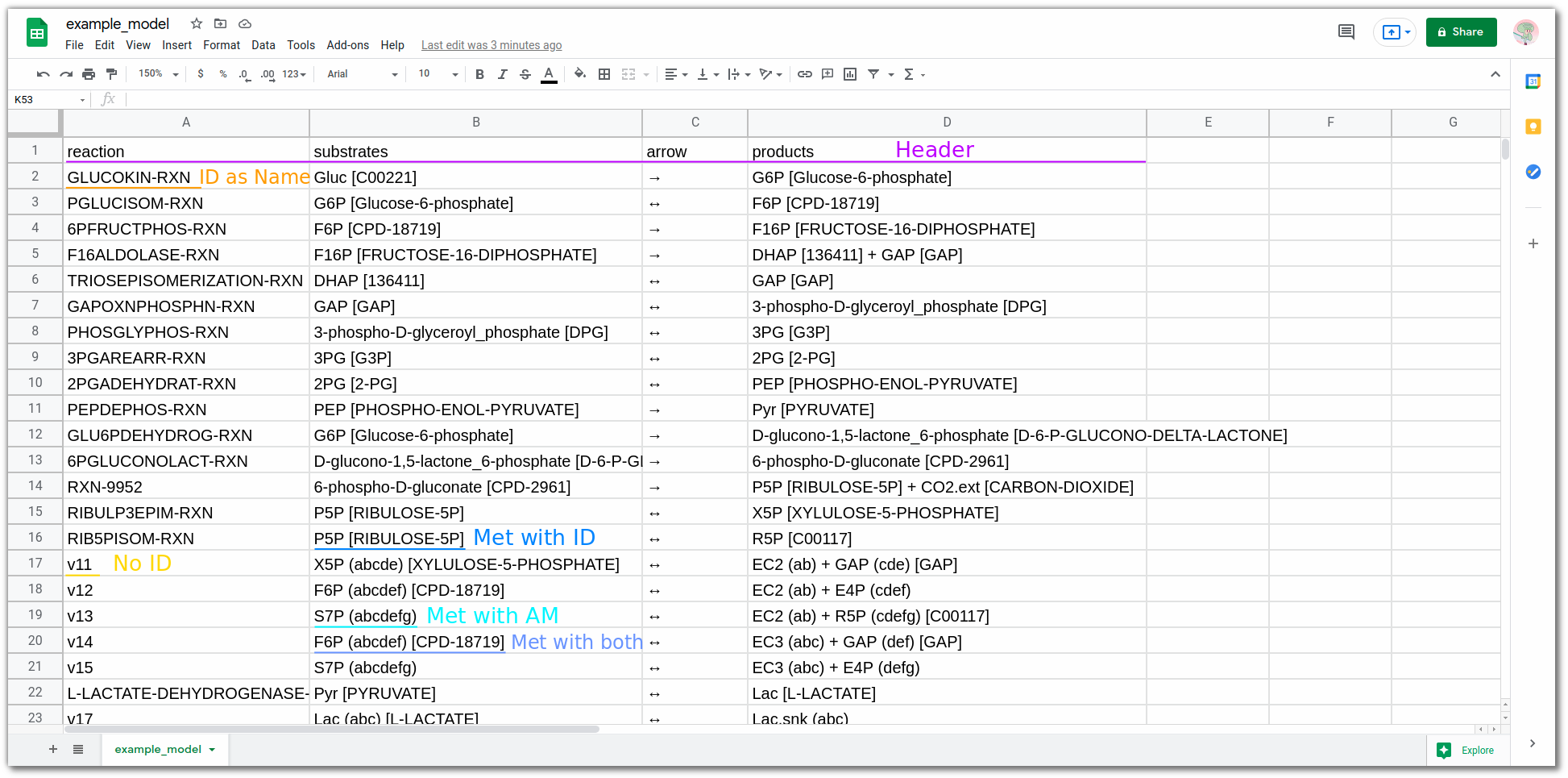
The file is structured in 4 columns:
Each column has special fields and options.
Reaction Name#
<ReactionName/Identifier> (<ReactionIdentifier>)
Reaction names can be freely chosen, but require a reaction identifier from either 🔗 BRENDA, 🔗 KEGG, or 🔗 MetaCyc to get a database atom mapping. Reaction_identifiers can either be used as the name itself or can be given in parenthesis/square brackets behind the name Name (Reaction_identifier) .
For example:
- Reaction1 (F16ALDOLASE-RXN) - Reaction identifier in parenthesis
- F16ALDOLASE-RXN - Reaction identifier as name
- Reaction3 - No identifier, no mapping
Substrates#
<MetaboliteName> (<AtomMapping>) [<MetaboliteIdentifier>] + <Metabolite...
Metabolites names, as in the reaction column, can be freely chosen and multiple metabolites are split with a plus sign +.
Metabolite Identifiers map metabolites to a given reaction and are required if a database atom mapping is wanted. To use metabolite identifiers square brackets can be used Acetyl-CoA [ACETYL-COA]. Metabolite identifiers from the above databases can be utilized (🔗 BRENDA, 🔗 KEGG, or 🔗 MetaCyc) and database specific identifiers can be mixed.
Optional parameters include Manual Atom Mappings, which can be used inside parenthesis Acetyl-CoA (bc) after the metabolite name. But why would you need manual atom mappings? Lets say you have a theoretical reaction that is not annotated in any database or you want to use a simplified reaction (see below). The manual atom mapping needs to be in the ABC-Format.

If a database reaction includes metabolites that should be removed from the atom mapping model the metabolite can be excluded by removing it from the model. Some applications could be the removal of cofactors or the simplification of the respective reaction.
For example:
- Acetyl-CoA (ab) + CO2 (c) - Metabolites with manual atom mappings
- Pyruvate [Pyr] + ... - Metabolite with metabolite identifier
Reaction Arrow#
The following reaction arrows can be used:
- Forward: -->, ->
- Reverse: <--, <-
- Reversible: <=>, <->
Products#
Products effectively function like Substrates.